Lab 13
Statistical Models
This lab
exercise is slightly different from the other ones involving ![]() . Remember that
. Remember that ![]() is a full-fledged statistics package for professional
applications and professionals often see their subject matter differently from
how it is taught in introductory courses. Correlation and regression are seen
by them as part of a larger concept of statistical models and we will have to
cover this concept before we can embark on applying
is a full-fledged statistics package for professional
applications and professionals often see their subject matter differently from
how it is taught in introductory courses. Correlation and regression are seen
by them as part of a larger concept of statistical models and we will have to
cover this concept before we can embark on applying ![]() routines. And the analysis of variance (ANOVA)
that we discussed in the lecture as part of sample-based procedures, turns out
to be a special kind of statistical model. Models and hypotheses succeed in
simplifying complex situations by ignoring information outside their frame and
by accurate generalization within it.
routines. And the analysis of variance (ANOVA)
that we discussed in the lecture as part of sample-based procedures, turns out
to be a special kind of statistical model. Models and hypotheses succeed in
simplifying complex situations by ignoring information outside their frame and
by accurate generalization within it.
Step 1 Deterministic models
Mathematical
or deterministic models are used to describe regular law-like behavior. In
fundamental research in the physical sciences, deterministic models are often
adequate. Statistical variability is so small that it can be ignored.
> x = 1:5
>
y = 0.5 * 9.8 * x^2
>
plot(y~x, xlab = “Time (s) “, ylab = “Distance fallen
(m) “)
> lines (spline(x,y)) # pass a smooth curve through the points
Step 2 Statistical models
Statistical
models rely on probabilistic forms of description that have wide application
over all areas of science. They often consist of a deterministic component as
well as a random component that attempts to account for variation that is not
accounted for by a law-like property. In engineering terms, the deterministic
part is called a signal and the
random part noise. Typically, we
assume that the elements of the noise component are uncorrelated. Take the roller.txt table for instance. Here,
different weights of roller were rolled over different parts of a lawn, and the
depression noted. We might expect depression to be proportional to roller
weight – the signal part. The variation exhibited in the values for depression/weight makes it clear that this is not
the whole story. We therefore model deviations from the line as random noise:
depression = b * weight + noise
Here b is
a constant, which we do not know but can try to estimate. The noise is different
for each different part of the lawn. If there were no noise, all the points
would lie exactly on a line.
> roller = read.table(“roller.txt”, header=T)
>
roller
>
plot(depression ~ weight, data=roller, xlab=”Weight of
roller (t) “, ylab=”Depression (mm) “)
> abline(0,2.25) # a slope of 2.25 looks about right
Many
models have the form
observed value = model prediction +
statistical error,
or
mathematically:
Y = m + e
Using the
mathematical idea of expected value, it is usual to define m = E(Y ). The
error term e is assumed
to be independent and normally distributed.
After
fitting a line through the lawn roller data, we can predict the depression that
would result for a given weight. The accuracy of the prediction depends upon
the amount of noise.
Step 3 Model formulae
![]() ’s
modeling functions use formulae to describe the role of variables in models. A
large part of the analyst’s task is to find the model formula that will be
effective for the task in hand. For example, the following model statement with
formula depression ~
weight, fits a line to
the roller data:
’s
modeling functions use formulae to describe the role of variables in models. A
large part of the analyst’s task is to find the model formula that will be
effective for the task in hand. For example, the following model statement with
formula depression ~
weight, fits a line to
the roller data:
> lm(depression ~
weight, data =roller) # prints
out summary information
The lm() function stands for linear model. Note that this
fits a model that has an intercept term yi
= a
+ bxi. The nonrandom part of the
equation describes yi as
lying on a straight line. The slope of the line (the regression coefficient) is b, the increase per unit change in x. The line intersects the y-axis at the intercept a. For a
model that omits the intercept term, specify
> lm(depression ~ -1 +
weight, data = roller)
The
parameters a and b can be
estimated using the method of least squares.
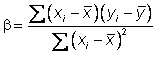
![]()
The
argument to lm() is a model formula, in which the tilde
symbol (~) should be read as “described by”. This has been seen several times
earlier, both in connection with boxplots and stripcharts and with the t
and Wilcoxon tests.
The lm() function handles much more complicated models than
simple linear regression. There can be many other things besides a dependent
and a descriptive variable in a model formula. In its raw form, the output of lm() is very brief. All you see is the estimated
intercept a and the estimated slope b.
Step 4 Model objects
The result
of lm() is a model object. This is a distinct concept of ![]() .
Where other statistical software packages focus on generating printed output,
you get instead the result of a model fit encapsulated in an object from which the desired
quantities can be obtained using extractor
functions. An lm object does in fact contain much
more information than you see when it is printed. A basic extractor function is
summary():
.
Where other statistical software packages focus on generating printed output,
you get instead the result of a model fit encapsulated in an object from which the desired
quantities can be obtained using extractor
functions. An lm object does in fact contain much
more information than you see when it is printed. A basic extractor function is
summary():
> summary(lm(depression~weight, data=roller))
The
following is a dissection of the output.
Call:
lm(formula
= depression ~ weight, data = roller)
Like in t.test(), etc., the output starts with
something that is essentially a repeat of the function call. This is not very
interesting when one has just given it as a command to ![]() ,
but it is useful if the result is saved in a variable that is printed later.
,
but it is useful if the result is saved in a variable that is printed later.
Residuals:
Min 1Q Median 3Q
Max
-8.180 -5.580 -1.346 5.920
8.020
This gives
a superficial view of the distribution of the residuals, which may be used to
check distributional assumptions. The average of the residuals is zero by
definition, so the median should not be far from zero and the minimum and the
maximum should be roughly equal in absolute value.
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -2.0871
4.7543 -0.439 0.67227
weight 2.6667 0.7002
3.808 0.00518 **
---
Signif. codes: 0 `***' 0.001 `**' 0.01 `*' 0.05 `.' 0.1 ` '
1
Here we
see the regression coefficient and the intercept again, but this time with accompanying
standard errors, t tests, and p –values. The symbols to the right are graphical indicators of the
level of significance. The line below the table shows the definition of these
indicators; one star means 0.01 < p
< 0.05.
Residual standard error: 6.735 on 8 degrees of
freedom
This is
the residual variation, an expression of the variation of the observation
around the regression line, estimating the model parameter s.
Multiple R-Squared: 0.6445, Adjusted R-squared: 0.6001
The first
term above is R2, which in
simple linear regression may be recognized as the squared Pearson correlation
coefficient. The other one is the adjusted R2;
if you multiply it by 100%, it can be interpreted as “% variance reduction”.
F-statistic:
14.5 on 1 and 8 DF, p-value:
0.005175
This is an
F test for the hypothesis that the
regression coefficient is zero. This test is not really interesting in a simple
linear regression analysis since it just duplicates information already given –
it becomes more interesting when there is more than one explanatory variable.
Notice that it gives the exact same result as the t test for a zero slope. In
fact, the F test is identical to the
square of the t test: 14.5 = (3.808)2. This is
true in any model with one degree of freedom.
Now we
don’t need to guess the regression line anymore as it fits best through the roller measurements. In
> plot(depression ~ weight, data=roller,
xlab=”Weight of roller (t) “, ylab=”Depression (mm) “)
> abline(lm(depression ~ weight,
data=roller))
We now use
the content of the lm object to define a line based on
the intercept (a) and slope (b) (hence the name of the function abline()). It can be used with scalar
values as in abline(1.1,
0.022), but
conveniently it can also extract the information from the linear model fitted
to data with lm().
Step 5 Residual and fitted values
We have
seen how summary() can be used to extract information
about the results of a regression analysis. Two further extraction functions
are fitted() and resid(). They are used as follows. For convenience, we first store the value
returned by lm() under the name lm.roller, but you could of course use any
other name.
> lm.roller
= lm(depression ~ weight, data=roller)
> fitted(lm.roller)
> resid(lm.roller)
The function
fitted() returns fitted values – the y-values that you would expect for given
x-values according to the
best-fitting straight line: in the present case -2.087 + 2.66 weight. The residuals shown by resid() is the difference between this and the observed depression.
Step 6 Prediction and confidence intervals
Fitted
lines are often presented with uncertainty bands around them. There are two
kinds of bands, often referred to as the “narrow” and “wide” limits. The narrow
bands, confidence bands, reflect the
uncertainty about the line itself. If there are many observations, the bands
will be quite narrow, reflecting a well-determined line.
The wide
bands, prediction bands, include
uncertainty about future observations. The bands should capture the majority of
observed points and will not collapse to a line as the number of observations
increases. Rather, the limits approach the true line ±2 standard deviations (for 95%
limits).
Predicted
values, with or without prediction and confidence bands, may be extracted with
the function predict(). With no arguments, it gives the
fitted values:
>
predict(lm.roller)
If you add
interval=”confidence” or “interval=”prediction” then you get the vector of predicted values
augmented with limits. The arguments can be abbreviated:
>
predict(lm.roller, int=”c”)
>
predict(lm.roller, int=”p”)
fit is
the expected values, here identical to the fitted values. lwr and upr (lower/upper) are the confidence limits for the expected values,
respectively, the prediction limits for depression with these limits of weight.
Step 7 Correlation
A
correlation coefficient is a measure of association between two random
variables. It ranges from -1 to +1, where the extremes indicate perfect
correlation and 0 means no correlation. The function cor() can be used to compute the correlation between two
or more vectors.
> cor(roller)
Of course,
this is more interesting when the data frame contains more than two vectors.
However, the above calculations give no indication of whether the correlation
is significantly different from zero. To that end, you need cor.test(). It works simply by specifying the
two variables:
> cor.test(roller$depression, roller$weight)
Notice that it is the same p-value
as in the regression analysis and also as that based on the ANOVA table for the
regression model that will be described in step 8.
As with
the one- and two-sample problems, you may be interested in nonparametric
variants. These have the advantage of not depending on the normal distribution.
The main disadvantage is that its interpretation is not quite clear. Unlike
group comparisons where there is essential one function per named test,
correlation tests are all grouped into cor.test(). There is no special spearman.test()
function. Instead, the test is considered one of several possibilities for
testing correlations and is therefore specified via an option to cor.test():
> cor.test(roller$depression, roller$weight, method=”spearman”)
Step 8 ANOVA
Simple
analysis of variance can be performed in ![]() using the function lm(). For more elaborate analyses, there is anova(). We calculate a model object using lm() and extract the analysis of variance table with anova().
using the function lm(). For more elaborate analyses, there is anova(). We calculate a model object using lm() and extract the analysis of variance table with anova().
> anova(lm(depression~weight, data =roller)
In the
lecture, I referred to “between groups” and “within groups” sums of squares. ![]() uses a slightly
different labeling. Variation between groups is labeled by the name of the
grouping factor (here weight) and variation within groups is
labeled Residual. ANOVA tables can be used for a wide
range of statistical models, and it is convenient to use a format that is less
linked to the particular problem of comparing groups.
uses a slightly
different labeling. Variation between groups is labeled by the name of the
grouping factor (here weight) and variation within groups is
labeled Residual. ANOVA tables can be used for a wide
range of statistical models, and it is convenient to use a format that is less
linked to the particular problem of comparing groups.
With this,
we are ready to turn you loose on trying yourself with statistical modeling.
Here are your tasks for today’s lab. As before, record all your answers to the
above six questions and send them in an ASCII email message to Jing Li.
![]() 1 Use read.csv() to import the homes.txt table that we discussed in the
lecture and calculate the ANOVA F test statistic. Do the four neighborhoods
represent one or more housing markets?
1 Use read.csv() to import the homes.txt table that we discussed in the
lecture and calculate the ANOVA F test statistic. Do the four neighborhoods
represent one or more housing markets?
![]() 2 Read
the snow.txt table that we discussed in the lecture and determine the
correlation coefficient r. What is the t test statistic or the p
value indicating the significance of the result?
2 Read
the snow.txt table that we discussed in the lecture and determine the
correlation coefficient r. What is the t test statistic or the p
value indicating the significance of the result?
![]() 3 What
is the regression line equation that best fits the snowfall data?
3 What
is the regression line equation that best fits the snowfall data?
![]() 4 How
much of the variation (in %) is explained by the equation that you calculated
in Q3?
4 How
much of the variation (in %) is explained by the equation that you calculated
in Q3?